CodelistGenerator
2025-07-06
Working with the OMOP CDM from R
Standarisation of the data format
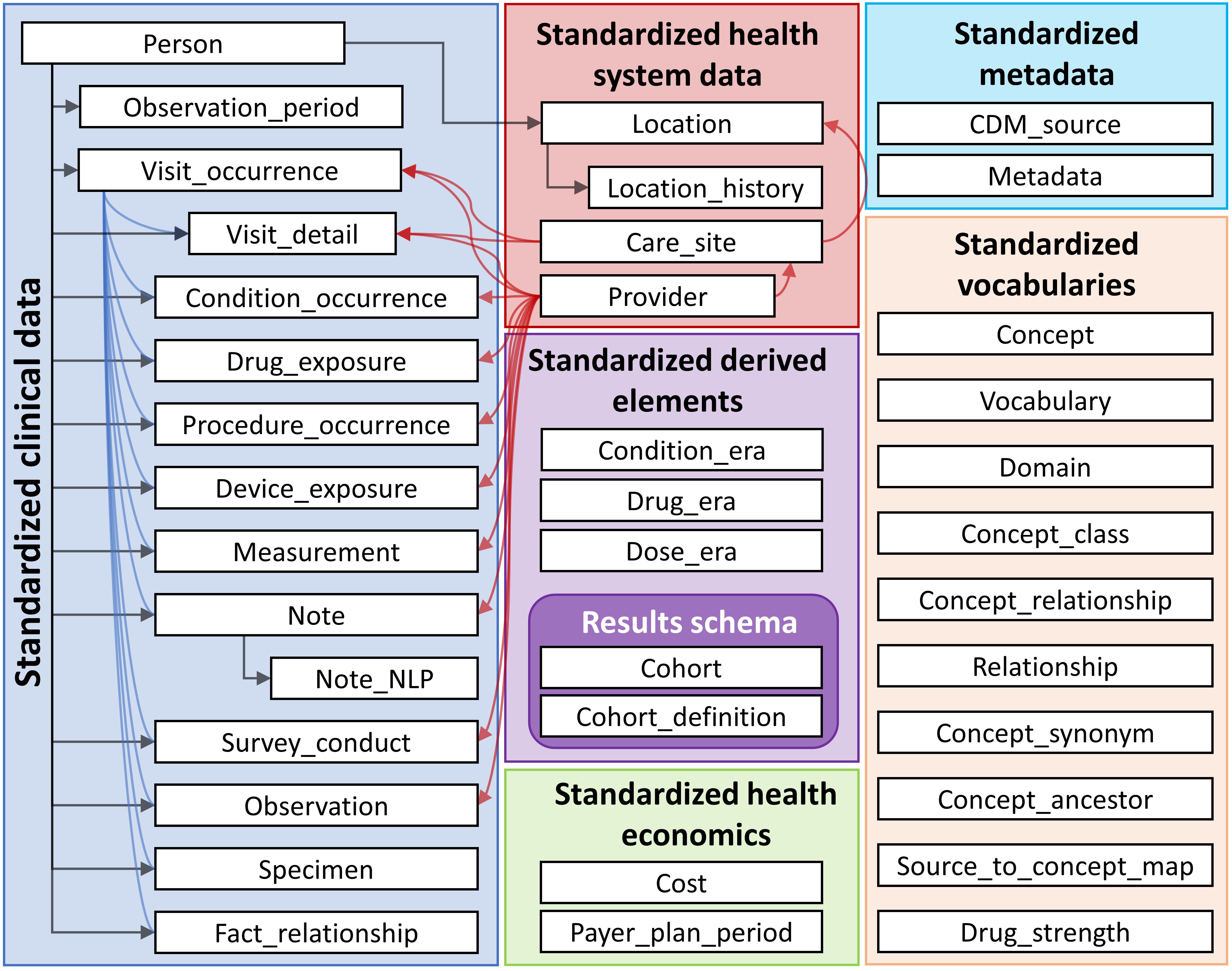
The OMOP Common Data Model
Create a reference to GiBleed dataset
Eunomia is a synthetic OMOP database with ~2,600 individuals. It is freely available and you can create a cdm reference like:
con <- dbConnect(duckdb(),
dbdir = eunomiaDir())
cdm <- cdmFromCon(con = con,
cdmSchema = "main",
writeSchema = "main",
cdmName = "Eunomia")
cdm── # OMOP CDM reference (duckdb) of Eunomia ────────────────────────────────────────────────────────────────────────────• omop tables: person, observation_period, visit_occurrence, visit_detail, condition_occurrence, drug_exposure,
procedure_occurrence, device_exposure, measurement, observation, death, note, note_nlp, specimen, fact_relationship,
location, care_site, provider, payer_plan_period, cost, drug_era, dose_era, condition_era, metadata, cdm_source,
concept, vocabulary, domain, concept_class, concept_relationship, relationship, concept_synonym, concept_ancestor,
source_to_concept_map, drug_strength• cohort tables: -• achilles tables: -• other tables: -Creating a reference to the OMOP common data model
Once we have created the our reference to the overall OMOP CDM, we can reference specific tables using the “$” operator or [[““]].
cdm$observation_period |> head(2)# Source: SQL [?? x 5]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
observation_period_id person_id observation_period_start_date observation_period_end_date period_type_concept_id
<int> <int> <date> <date> <int>
1 6 6 1963-12-31 2007-02-06 44814724
2 13 13 2009-04-26 2019-04-14 44814724cdm[["observation_period"]] |> head(2)# Source: SQL [?? x 5]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
observation_period_id person_id observation_period_start_date observation_period_end_date period_type_concept_id
<int> <int> <date> <date> <int>
1 6 6 1963-12-31 2007-02-06 44814724
2 13 13 2009-04-26 2019-04-14 44814724Behind the scenes
The omopgenerics package defines core classes and methods used by CDMConnector and analytic packages.
Having omopgenerics as a central dependency reduces code duplication and ensures consistency across packages (eg function inputs, error messages, and results objects).
omopgenerics::validateCdmArgument(cdm = cdm,
checkOverlapObservation = TRUE,
checkStartBeforeEndObservation = TRUE,
checkPlausibleObservationDates = TRUE)
<cdm_reference> object
class(cdm)[1] "cdm_reference"cdmName(cdm)[1] "Eunomia"cdmVersion(cdm)[1] "5.3"cdmSource(cdm)This is a duckdb cdm source
<cdm_table> object
cdm$person# Source: table<person> [?? x 18]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
person_id gender_concept_id year_of_birth month_of_birth day_of_birth birth_datetime race_concept_id
<int> <int> <int> <int> <int> <dttm> <int>
1 6 8532 1963 12 31 1963-12-31 00:00:00 8516
2 123 8507 1950 4 12 1950-04-12 00:00:00 8527
3 129 8507 1974 10 7 1974-10-07 00:00:00 8527
4 16 8532 1971 10 13 1971-10-13 00:00:00 8527
5 65 8532 1967 3 31 1967-03-31 00:00:00 8516
6 74 8532 1972 1 5 1972-01-05 00:00:00 8527
7 42 8532 1909 11 2 1909-11-02 00:00:00 8527
8 187 8507 1945 7 23 1945-07-23 00:00:00 8527
9 18 8532 1965 11 17 1965-11-17 00:00:00 8527
10 111 8532 1975 5 2 1975-05-02 00:00:00 8527
# ℹ more rows
# ℹ 11 more variables: ethnicity_concept_id <int>, location_id <int>, provider_id <int>, care_site_id <int>,
# person_source_value <chr>, gender_source_value <chr>, gender_source_concept_id <int>, race_source_value <chr>,
# race_source_concept_id <int>, ethnicity_source_value <chr>, ethnicity_source_concept_id <int>class(cdm$person)[1] "omop_table" "cdm_table" "tbl_duckdb_connection" "tbl_dbi"
[5] "tbl_sql" "tbl_lazy" "tbl"
<cdm_table> object
cdmReference(cdm$person)── # OMOP CDM reference (duckdb) of Eunomia ────────────────────────────────────────────────────────────────────────────• omop tables: person, observation_period, visit_occurrence, visit_detail, condition_occurrence, drug_exposure,
procedure_occurrence, device_exposure, measurement, observation, death, note, note_nlp, specimen, fact_relationship,
location, care_site, provider, payer_plan_period, cost, drug_era, dose_era, condition_era, metadata, cdm_source,
concept, vocabulary, domain, concept_class, concept_relationship, relationship, concept_synonym, concept_ancestor,
source_to_concept_map, drug_strength• cohort tables: -• achilles tables: -• other tables: -tableName(cdm$person)[1] "person"tableSource(cdm$person)This is a duckdb cdm source
<cdm_table> object
cdmName(cdm$person)[1] "Eunomia"cdmVersion(cdm$person)[1] "5.3"cdmSource(cdm$person)This is a duckdb cdm sourceWorking with the cdm reference
We can use common dplyr operations to interact with the data in our cdm reference
cdm$condition_occurrence |>
count()# Source: SQL [?? x 1]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
n
<dbl>
1 65332cdm$condition_occurrence |>
summarise(
min_condition_start = min(condition_start_date, na.rm = TRUE),
median_condition_start = median(condition_start_date, na.rm = TRUE),
max_condition_start = max(condition_start_date, na.rm = TRUE))# Source: SQL [?? x 3]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
min_condition_start median_condition_start max_condition_start
<date> <dttm> <date>
1 1908-10-30 1986-12-09 00:00:00 2019-06-29 Dplyr R code to SQL
Behind the scenes our dplyr query is being translated into SQL.
cdm$condition_occurrence |>
count() |>
show_query()<SQL>
SELECT COUNT(*) AS n
FROM condition_occurrencecdm$condition_occurrence |>
summarise(
min_condition_start = min(condition_start_date, na.rm = TRUE),
median_condition_start = median(condition_start_date, na.rm = TRUE),
max_condition_start = max(condition_start_date, na.rm = TRUE)) |>
show_query()<SQL>
SELECT
MIN(condition_start_date) AS min_condition_start,
MEDIAN(condition_start_date) AS median_condition_start,
MAX(condition_start_date) AS max_condition_start
FROM condition_occurrenceYour turn
Using a cdm reference you have connected to:
How many people are in the person table?
What is the minimum observation period start date?
What is the maximum observation period end date?
Solution
💡 Click to see solution
cdm$person |>
count()# Source: SQL [?? x 1]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
n
<dbl>
1 2694cdm$observation_period |>
summarise(min_obs_period_start = min(observation_period_start_date, na.rm = TRUE)) # Source: SQL [?? x 1]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
min_obs_period_start
<date>
1 1908-09-22 cdm$observation_period |>
summarise(max_obs_period_start = max(observation_period_end_date, na.rm = TRUE))# Source: SQL [?? x 1]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
max_obs_period_start
<date>
1 2019-07-03 CDM vocabulary tables
CDM vocabulary tables

CDM vocabulary tables
cdm$concept |> glimpse()Rows: ??
Columns: 10
Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
$ concept_id <int> 35208414, 1118088, 40213201, 1557272, 4336464, 4295880, 3020630, 19129655, 44923712, 1569708,…
$ concept_name <chr> "Gastrointestinal hemorrhage, unspecified", "celecoxib 200 MG Oral Capsule [Celebrex]", "pneu…
$ domain_id <chr> "Condition", "Drug", "Drug", "Drug", "Procedure", "Procedure", "Measurement", "Drug", "Drug",…
$ vocabulary_id <chr> "ICD10CM", "RxNorm", "CVX", "RxNorm", "SNOMED", "SNOMED", "LOINC", "RxNorm", "NDC", "ICD10CM"…
$ concept_class_id <chr> "4-char billing code", "Branded Drug", "CVX", "Ingredient", "Procedure", "Procedure", "Lab Te…
$ standard_concept <chr> NA, "S", "S", "S", "S", "S", "S", "S", NA, NA, "S", "S", "S", "S", "S", "S", NA, "S", "S", "S…
$ concept_code <chr> "K92.2", "213469", "33", "46041", "232717009", "76601001", "2885-2", "789980", "00025152531",…
$ valid_start_date <date> 2007-01-01, 1970-01-01, 2008-12-01, 1970-01-01, 1970-01-01, 1970-01-01, 1970-01-01, 2008-03-…
$ valid_end_date <date> 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-…
$ invalid_reason <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…CDM vocabulary tables
cdm$condition_occurrence |>
group_by(condition_concept_id) |>
tally() |>
left_join(
cdm$concept |>
select("concept_id", "concept_name"),
by = c("condition_concept_id" = "concept_id")
) |>
collect() |>
arrange(desc(n))# A tibble: 80 × 3
condition_concept_id n concept_name
<int> <dbl> <chr>
1 40481087 17268 Viral sinusitis
2 4112343 10217 Acute viral pharyngitis
3 260139 8184 Acute bronchitis
4 372328 3605 Otitis media
5 80180 2694 Osteoarthritis
6 28060 2656 Streptococcal sore throat
7 81151 1915 Sprain of ankle
8 378001 1013 Concussion with no loss of consciousness
9 4283893 1001 Sinusitis
10 4294548 939 Acute bacterial sinusitis
# ℹ 70 more rowsCDM vocabulary tables
cdm$concept_ancestor |> glimpse()Rows: ??
Columns: 4
Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
$ ancestor_concept_id <int> 4180628, 4179141, 21500574, 21505770, 21503967, 36203060, 36151386, 21502552, 4076562…
$ descendant_concept_id <int> 313217, 4146173, 1118084, 1119510, 40162522, 40479422, 1119510, 1112807, 40769189, 19…
$ min_levels_of_separation <int> 5, 2, 4, 0, 5, 4, 0, 4, 2, 2, 0, 0, 0, 0, 0, 0, 0, 0, 2, 0, 0, 2, 2, 0, 0, 2, 0, 0, 0…
$ max_levels_of_separation <int> 6, 2, 4, 0, 6, 4, 0, 4, 2, 2, 0, 0, 0, 0, 0, 0, 0, 0, 3, 0, 0, 3, 3, 0, 0, 3, 0, 0, 0…CDM vocabulary tables
cdm$concept_relationship |> glimpse()Rows: ??
Columns: 6
Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
$ concept_id_1 <int> 192671, 1118088, 1569708, 35208414, 35208414, 40162359, 44923712, 45011828
$ concept_id_2 <int> 35208414, 44923712, 35208414, 192671, 1569708, 45011828, 1118088, 40162359
$ relationship_id <chr> "Mapped from", "Mapped from", "Subsumes", "Maps to", "Is a", "Mapped from", "Maps to", "Maps …
$ valid_start_date <date> 1970-01-01, 1970-01-01, 2016-03-25, 1970-01-01, 2016-03-25, 2009-08-03, 1970-01-01, 2009-08-0…
$ valid_end_date <date> 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-3…
$ invalid_reason <chr> NA, NA, NA, NA, NA, NA, NA, NACDM vocabulary tables
cdm$concept_synonym |> glimpse()Rows: ??
Columns: 3
Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
$ concept_id <int> 964261, 1322184, 441267, 1718412, 4336464, 4102123, 4237458, 4280726, 4330583, 3014576, 4…
$ concept_synonym_name <chr> "cyanocobalamin 5000 MCG/ML Injectable Solution", "clopidogrel", "Cystic fibrosis (disord…
$ language_concept_id <int> 4180186, 4180186, 4180186, 4180186, 4180186, 4180186, 4180186, 4180186, 4180186, 4180186,…Exploring vocabulary tables using CodelistGenerator
Vocabulary version
Search results will be specific to the version of the vocabulary being used
getVocabVersion(cdm = cdm)[1] "v5.0 18-JAN-19"Available vocabularies
What vocabularies are available?
getVocabularies(cdm = cdm)[1] "CVX" "Gender" "ICD10CM" "LOINC" "NDC" "None" "RxNorm" "SNOMED" "Visit" Available domains
What domains are present?
getDomains(cdm = cdm)[1] "Drug" "Measurement" "Procedure" "Condition" "Observation" "Visit" "Gender" Concept classes
What concept classes are present?
getConceptClassId(
cdm = cdm,
standardConcept = "Standard",
domain = "Drug"
)[1] "Branded Drug" "Branded Drug Comp" "Branded Pack" "Clinical Drug" "Clinical Drug Comp"
[6] "CVX" "Ingredient" "Quant Branded Drug" "Quant Clinical Drug"getConceptClassId(
cdm = cdm,
standardConcept = "Standard",
domain = "Condition"
)[1] "Clinical Finding"Your turn
Using a cdm reference you have connected to:
What is the vocabulary version of the cdm?
How many concepts are in your concept table?
What domains are available?
Solution
💡 Click to see solution
getVocabVersion(cdm = cdm)[1] "v5.0 18-JAN-19"cdm$concept |>
tally()# Source: SQL [?? x 1]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
n
<dbl>
1 444getDomains(cdm = cdm)[1] "Drug" "Procedure" "Condition" "Measurement" "Observation" "Visit" "Gender" Vocabulary based codelists using CodelistGenerator
Vocabulary-based codelists using CodelistGenerator
We can use drug hierarchies and relationships to create vocabulary-based codelists.
Drug ingredients
ingredients <- getDrugIngredientCodes(cdm = cdm,
name = c("warfarin", "morphine",
"verapamil", "atorvastatin",
"nitroglycerin", "simvastatin",
"acetaminophen"),
nameStyle = "{concept_name}")
ingredients
- acetaminophen (7 codes)
- atorvastatin (2 codes)
- morphine (2 codes)
- nitroglycerin (2 codes)
- simvastatin (2 codes)
- verapamil (2 codes)
along with 1 more codelistsingredients$warfarin[1] 1310149 40163554# Source: SQL [?? x 10]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
concept_id concept_name domain_id vocabulary_id concept_class_id standard_concept concept_code valid_start_date
<int> <chr> <chr> <chr> <chr> <chr> <chr> <date>
1 1310149 Warfarin Drug RxNorm Ingredient S 11289 1970-01-01
2 40163554 Warfarin Sodium 5 … Drug RxNorm Clinical Drug S 855332 2009-08-02
# ℹ 2 more variables: valid_end_date <date>, invalid_reason <chr>Drug ingredients
combinations <- getDrugIngredientCodes(cdm = cdm,
name = c("acetaminophen"),
ingredientRange = c(2, Inf),
nameStyle = "{concept_name}")
combinations
- acetaminophen (4 codes)combinations$acetaminophen[1] 40229134 40231925 40162522 19133768cdm$concept |>
filter(concept_id %in% c(40229134, 40231925, 40162522, 19133768)) |>
pull(concept_name)[1] "Acetaminophen 21.7 MG/ML / Dextromethorphan Hydrobromide 1 MG/ML / doxylamine succinate 0.417 MG/ML Oral Solution"
[2] "Acetaminophen 325 MG / Oxycodone Hydrochloride 5 MG Oral Tablet"
[3] "Acetaminophen 325 MG / Hydrocodone Bitartrate 7.5 MG Oral Tablet"
[4] "Acetaminophen 750 MG / Hydrocodone Bitartrate 7.5 MG Oral Tablet" Your turn
Using Eunomia data:
Get codes for memantine using
getDrugIngredientCodes. How many codes do you include?Get codes for memantine, restricting to only concepts with it as the only ingredient. Do the codes you include change?
Solution
💡 Click to see solution
memantine_codes <- getDrugIngredientCodes(cdm = cdm, name = "memantine")
memantine_codes
- 6719_memantine (3 codes)memantine_monotherapy_codes <- getDrugIngredientCodes(cdm = cdm, name = "memantine",
ingredientRange = c(1, 1))
memantine_monotherapy_codes
- 6719_memantine (2 codes)Systematic search using CodelistGenerator
Systematic search using CodelistGenerator
CodelistGenerator is used to create a candidate set of codes for helping to define patient cohorts in data mapped to the OMOP common data model.
A little like the process for a systematic review, the idea is that for a specified search strategy, CodelistGenerator will identify a set of concepts that may be relevant, with these then being screened to remove any irrelevant codes.
Codes for asthma
asthma_codes <- getCandidateCodes(
cdm = cdm,
keywords = "asthma",
domains = "Condition"
)
asthma_codes |> glimpse()Rows: 2
Columns: 6
$ concept_id <int> 4051466, 317009
$ found_from <chr> "From initial search", "From initial search"
$ concept_name <chr> "Childhood asthma", "Asthma"
$ domain_id <chr> "Condition", "Condition"
$ vocabulary_id <chr> "SNOMED", "SNOMED"
$ standard_concept <chr> "S", "S"asthma_cs <- newCodelist(list("asthma" = asthma_codes$concept_id))
asthma_cs
- asthma (2 codes)Your turn
Using Eunomia data:
Search for codes for sinusitis recorded in the condition domain
Do you identify any more codes if you also search in the observation domain as well as the condition domain
Solution
💡 Click to see solution
sinusitis_codes <- getCandidateCodes(
cdm = cdm,
keywords = "sinusitis",
domains = "Condition"
)
nrow(sinusitis_codes)[1] 4sinusitis_codes_2 <- getCandidateCodes(
cdm = cdm,
keywords = "sinusitis",
domains = c("Condition", "Observation")
)
nrow(sinusitis_codes_2)[1] 4Codelists to cohorts
Codelists to cohorts
medication_codes <- getDrugIngredientCodes(cdm = cdm,
name = c("warfarin", "morphine",
"verapamil", "atorvastatin",
"nitroglycerin", "simvastatin",
"acetaminophen"),
nameStyle = "{concept_name}")
cdm$meds <- conceptCohort(cdm = cdm,
conceptSet = medication_codes,
exit = "event_end_date",
name = "meds") |>
collapseCohorts(gap = 30)
cdm$meds # Source: table<meds> [?? x 4]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
cohort_definition_id subject_id cohort_start_date cohort_end_date
<int> <int> <date> <date>
1 1 74 1973-09-28 1973-10-12
2 1 129 1994-08-25 1994-09-08
3 1 220 1990-01-12 1990-01-26
4 1 452 1965-12-31 1966-01-14
5 1 669 1998-06-15 1998-07-06
6 1 1091 1998-07-27 1998-08-10
7 1 1134 1955-06-08 1955-06-22
8 1 1135 1981-10-27 1981-11-26
9 1 1144 1972-06-09 1972-06-23
10 1 1144 1987-10-04 1987-10-11
# ℹ more rowsCodelists to cohorts
sinusitis_codes <- getCandidateCodes(
cdm = cdm,
keywords = "sinusitis",
domains = "Condition"
)
cdm$sinusitis <- conceptCohort(cdm = cdm,
conceptSet = list(sinusitis = sinusitis_codes$concept_id),
exit = "event_end_date",
name = "sinusitis") |>
requireIsFirstEntry()
cdm$sinusitis# Source: table<sinusitis> [?? x 4]
# Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
cohort_definition_id subject_id cohort_start_date cohort_end_date
<int> <int> <date> <date>
1 1 1 1967-05-30 1967-06-20
2 1 23 1991-08-08 1991-08-22
3 1 69 1952-04-20 1952-10-19
4 1 74 1979-11-30 1979-12-14
5 1 81 1956-08-27 1956-09-03
6 1 188 1967-08-03 1967-08-10
7 1 189 1972-06-02 1972-06-09
8 1 213 1968-12-30 1969-01-06
9 1 219 1973-02-09 1973-02-16
10 1 227 1972-03-20 1972-03-27
# ℹ more rows
<cohort_table> object
settings(cdm$meds)# A tibble: 7 × 4
cohort_definition_id cohort_name cdm_version vocabulary_version
<int> <chr> <chr> <chr>
1 1 acetaminophen 5.3 v5.0 18-JAN-19
2 2 atorvastatin 5.3 v5.0 18-JAN-19
3 3 morphine 5.3 v5.0 18-JAN-19
4 4 nitroglycerin 5.3 v5.0 18-JAN-19
5 5 simvastatin 5.3 v5.0 18-JAN-19
6 6 verapamil 5.3 v5.0 18-JAN-19
7 7 warfarin 5.3 v5.0 18-JAN-19 settings(cdm$sinusitis)# A tibble: 1 × 4
cohort_definition_id cohort_name cdm_version vocabulary_version
<int> <chr> <chr> <chr>
1 1 sinusitis 5.3 v5.0 18-JAN-19
<cohort_table> object
cohortCount(cdm$meds)# A tibble: 7 × 3
cohort_definition_id number_records number_subjects
<int> <int> <int>
1 1 13860 2679
2 2 57 54
3 3 35 35
4 4 208 192
5 5 182 182
6 6 137 137
7 7 137 137cohortCount(cdm$sinusitis)# A tibble: 1 × 3
cohort_definition_id number_records number_subjects
<int> <int> <int>
1 1 2689 2689
<cohort_table> object
attrition(cdm$meds)# A tibble: 49 × 7
cohort_definition_id number_records number_subjects reason_id reason excluded_records excluded_subjects
<int> <int> <int> <int> <chr> <int> <int>
1 1 14205 2679 1 Initial qualifying … 0 0
2 1 14205 2679 2 Record start <= rec… 0 0
3 1 14205 2679 3 Record in observati… 0 0
4 1 14205 2679 4 Non-missing sex 0 0
5 1 14205 2679 5 Non-missing year of… 0 0
6 1 13908 2679 6 Merge overlapping r… 297 0
7 1 13860 2679 7 Collapse cohort wit… 48 0
8 2 57 54 1 Initial qualifying … 0 0
9 2 57 54 2 Record start <= rec… 0 0
10 2 57 54 3 Record in observati… 0 0
# ℹ 39 more rowsattrition(cdm$sinusitis)# A tibble: 7 × 7
cohort_definition_id number_records number_subjects reason_id reason excluded_records excluded_subjects
<int> <int> <int> <int> <chr> <int> <int>
1 1 20033 2689 1 Initial qualifying e… 0 0
2 1 20033 2689 2 Record start <= reco… 0 0
3 1 20033 2689 3 Record in observation 0 0
4 1 20033 2689 4 Non-missing sex 0 0
5 1 20033 2689 5 Non-missing year of … 0 0
6 1 19147 2689 6 Merge overlapping re… 886 0
7 1 2689 2689 7 Restricted to first … 16458 0
<cohort_table> object
cohortCodelist(cdm$meds, 1)
- acetaminophen (7 codes)cohortCodelist(cdm$sinusitis, 1)
- sinusitis (4 codes)Your turn
Using Eunomia data:
- Create a cohort of people taking aspirin, collapse records less than 30 days, and require is first ever exposure
Solution
💡 Click to see solution
aspirin_codes <- getDrugIngredientCodes(cdm = cdm, name = "aspirin")
cdm$aspirin_cohort <- conceptCohort(cdm = cdm, conceptSet = aspirin_codes,
name = "aspirin_cohort") |>
collapseCohorts(gap = 30) |>
requireIsFirstEntry()
cdm$aspirin_cohort |>
glimpse()Rows: ??
Columns: 4
Database: DuckDB v1.3.1 [unknown@Linux 6.11.0-1015-azure:R 4.5.1//tmp/RtmpkGLTdC/file2a5f12d5ca24.duckdb]
$ cohort_definition_id <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ subject_id <int> 1, 69, 81, 189, 227, 244, 246, 254, 310, 332, 352, 362, 388, 432, 450, 480, 505, 580, 616…
$ cohort_start_date <date> 1970-12-03, 1947-11-05, 1963-09-11, 1970-01-08, 1974-04-04, 1926-05-07, 1961-11-15, 1992…
$ cohort_end_date <date> 1970-12-31, 1947-11-19, 1963-12-10, 1970-02-07, 1974-04-18, 1926-05-21, 1962-01-14, 1992…