library(omock)
library(CohortConstructor)
library(CohortCharacteristics)
library(dplyr)
cdm <- mockCdmFromDataset(datasetName = "synthea-covid19-10k", source = "duckdb")8 Adding cohorts to the CDM
8.1 What is a cohort?
When performing research with the OMOP CDM we often want to identify groups of individuals who share some set of characteristics. The criteria for including individuals can range from the seemingly simple (e.g. people diagnosed with asthma) to the much more complicated (e.g. adults diagnosed with asthma who had a year of prior observation time in the database prior to their diagnosis, had no prior history of chronic obstructive pulmonary disease, and no history of use of short-acting beta-antagonists).
The set of people we identify are cohorts, and the OMOP CDM has a specific structure by which they can be represented, with a cohort table having four required fields:
Cohort definition id a unique identifier for each cohort (multiple cohorts can be defined in the same cohort table)
Subject id a foreign key to the subject in the cohort - typically referring to records in the person table
Cohort start date date that indicates the start date of the record.
Cohort end date date that indicates the end date of the record.
Individuals must be defined in the person table and have an ongoing record in the observation period table to be part of a cohort. Individuals can enter a cohort multiple times, but the time periods in which they are in the cohort cannot overlap.
It is beyond the scope of this book to describe all the different ways cohorts could be created, however in this chapter we provide a summary of some of the key building blocks for cohort creation. Cohort-building pipelines can be created following these principles to create a wide range of study cohorts.
8.2 Set up
We’ll use our the same COVID-19 synthetic dataset that we used before for demonstrating how cohorts can be constructed.
8.3 General concept based cohort
Often study cohorts will be based around a specific clinical event identified by some set of clinical codes. Here, for example, we use the CohortConstructor package to create a cohort of people with Covid-19. For this we are identifying any clinical records with the code 37311061.
cdm$covid <- conceptCohort(cdm = cdm,
conceptSet = list("covid" = 37311061),
name = "covid")
cdm$covid# Source: table<results.test_covid> [?? x 4]
# Database: DuckDB 1.4.0 [unknown@Linux 6.11.0-1018-azure:R 4.4.1//tmp/Rtmp5e21wo/file38b330d37a2c.duckdb]
cohort_definition_id subject_id cohort_start_date cohort_end_date
<int> <int> <date> <date>
1 1 250 2021-01-02 2021-01-18
2 1 394 2020-04-09 2020-05-16
3 1 6755 2020-04-09 2020-05-15
4 1 7173 2020-12-02 2021-01-01
5 1 10197 2020-05-16 2020-06-04
6 1 5780 2020-11-20 2020-12-14
7 1 6601 2020-06-07 2020-06-23
8 1 7616 2020-07-30 2020-08-14
9 1 7837 2021-01-31 2021-02-21
10 1 3226 2020-06-14 2020-07-11
# ℹ more rowsNote that the name argument determines the name of the permanent table written in the database and as we have seen before we have have to be consistent assigning the tables to the cdm object, that’s my we used name = "covid" and then we were able to assign it to cdm$covid. Otherwise see this failing example:
cdm$not_covid <- conceptCohort(cdm = cdm,
conceptSet = list("covid" = 37311061),
name = "covid")Warning: ! `codelist` casted to integers.ℹ Subsetting table condition_occurrence using 1 concept with domain: condition.
ℹ Combining tables.
ℹ Creating cohort attributes.
ℹ Applying cohort requirements.
ℹ Merging overlapping records.
✔ Cohort covid created.Error in `[[<-`:
✖ You can't assign a table named covid to not_covid.
ℹ You can change the name using compute:
cdm[['not_covid']] <- yourObject |>
dplyr::compute(name = 'not_covid')
ℹ You can also change the name using the `name` argument in your function:
`name = 'not_covid'`.In the defining the cohorts above we have needed to provide concept IDs to define our cohort. But, where do these come from?
We can search for codes of interest using the CodelistGenerator package. This can be done using a text search with the function getCandidateCodes(). For example, we can have found the code we used above (and many others) like so:
library(CodelistGenerator)
getCandidateCodes(cdm = cdm,
keywords = c("coronavirus", "covid"),
domains = "condition",
includeDescendants = TRUE)Limiting to domains of interest
Getting concepts to include
Adding descendants
Search completed. Finishing up.
✔ 37 candidate concepts identified
Time taken: 0 minutes and 1 seconds# A tibble: 37 × 6
concept_id found_from concept_name domain_id vocabulary_id standard_concept
<int> <chr> <chr> <chr> <chr> <chr>
1 3656667 From initia… Cardiomyopa… Condition SNOMED S
2 3661885 From initia… Fever cause… Condition SNOMED S
3 37310286 From initia… Infection o… Condition SNOMED S
4 703447 From initia… High risk c… Condition SNOMED S
5 3661631 From initia… Lymphocytop… Condition SNOMED S
6 3661632 From initia… Thrombocyto… Condition SNOMED S
7 3655973 From initia… At increase… Condition SNOMED S
8 3661406 From initia… Acute respi… Condition SNOMED S
9 37016927 From initia… Pneumonia c… Condition SNOMED S
10 1340294 From initia… Exacerbatio… Condition OMOP Extensi… S
# ℹ 27 more rowsWe can also do automated searches that make use of the hierarchies in the vocabularies. Here, for example, we find the code for the drug ingredient Acetaminophen and all of it’s descendants.
getDrugIngredientCodes(cdm = cdm, name = "acetaminophen")── 1 codelist ──────────────────────────────────────────────────────────────────
- 161_acetaminophen (25747 codes)Note that in practice clinical expertise is vital in the identification of appropriate codes so as to decide which the codes are in line with the clinical idea at hand.
We can see that as well as having the cohort entries above, our cohort table is associated with several attributes.
First, we can see the settings associated with cohort.
settings(cdm$covid) |>
glimpse()Rows: 1
Columns: 4
$ cohort_definition_id <int> 1
$ cohort_name <chr> "covid"
$ cdm_version <chr> "5.3"
$ vocabulary_version <chr> "v5.0 22-JUN-22"In settings we can see the cohort name that by default is the name of the codelist used, in this case ‘covid’ as we used conceptSet = list(covid = 37311061). Also the cdm and vocabulary versions are recorded in the settings by the CohortConstructor package.
Second, we can get counts of each cohort.
cohortCount(cdm$covid) |>
glimpse()Rows: 1
Columns: 3
$ cohort_definition_id <int> 1
$ number_records <int> 964
$ number_subjects <int> 964Where you can see the number of records and number of subjects for each cohort, in this case there are no multiple records per subject.
An attrition can also be retrieved from any cohort.
attrition(cdm$covid) |>
glimpse()Rows: 6
Columns: 7
$ cohort_definition_id <int> 1, 1, 1, 1, 1, 1
$ number_records <int> 964, 964, 964, 964, 964, 964
$ number_subjects <int> 964, 964, 964, 964, 964, 964
$ reason_id <int> 1, 2, 3, 4, 5, 6
$ reason <chr> "Initial qualifying events", "Record in observati…
$ excluded_records <int> 0, 0, 0, 0, 0, 0
$ excluded_subjects <int> 0, 0, 0, 0, 0, 0And finally, you can extract the codelists used to create a cohort table:
codelist <- cohortCodelist(cdm$covid, cohortId = 1)
codelist── 1 codelist ──────────────────────────────────────────────────────────────────
- covid (1 codes)codelist$covid[1] 37311061Note in this case we had to provide the cohortId of the cohort of interest.
All these attributes can be retrieved because it is a cohort_table object, a class on top of the usual cdm_table class that we have seen before:
class(cdm$covid)[1] "cohort_table" "cdm_table" "GeneratedCohortSet"
[4] "tbl_duckdb_connection" "tbl_dbi" "tbl_sql"
[7] "tbl_lazy" "tbl" As we will see below these attributes of the cohorts become particularly useful as we apply further restrictions on our cohort.
All these attributes that we have seen are part of the attributes of the cohort_table object and are used by these utility functions:
names(attributes(cdm$covid))[1] "names" "class" "tbl_source" "tbl_name"
[5] "cohort_set" "cohort_attrition" "cohort_codelist" "cdm_reference" In particular the cohort_set (contains the settings() source), cohort_attrition (contains the source for cohortCount() and attrition()) and cohort_codelist (contains the source for cohortCodelist()) attributes are the ones of interest. For database back-ends these attributes are stored in the database so when we read them again the attributes persist. See that even apparently we only have one table ‘cdm$covid’ in fact four tables were written in the database:
library(omopgenerics) # TODO https://github.com/OHDSI/omock/issues/189
Attaching package: 'omopgenerics'The following object is masked from 'package:stats':
filterlistSourceTables(cdm = cdm)[1] "covid" "covid_attrition" "covid_codelist" "covid_set" We do not have to worry about the attributes and the naming of the tables as CohortConstructor, CDMConnector and omopgenerics take care of that and if we create the cohorts with functions such as conceptCohort() then we will be able to read them back with the cohortTables argument of cdmFromCon() or the readSourceTable() function and all the attributes will be in place.
8.4 Applying inclusion criteria
8.4.1 Only include first cohort entry per person
Let’s say we first want to restrict to first entry.
cdm$covid <- cdm$covid |>
requireIsFirstEntry() 8.4.2 Restrict to study period
Then we are interested in records only after January 1st 2020.
cdm$covid <- cdm$covid |>
requireInDateRange(dateRange = c(as.Date("2020-09-01"), NA))8.4.3 Applying demographic inclusion criteria
Finally we want to restrict our population of interest to only adult males under 65 years old. We can do that with the requireDemographics() function.
cdm$covid <- cdm$covid |>
requireDemographics(ageRange = c(18, 64), sex = "Male")Note that all these require*() functions that come from the CohortConstructor package use functionalities from PatientProfiles and the naming is consistent, for example requireDemographics() uses addDemographics(), requirePriorObservation() uses addPriorObservation() and so…
8.4.4 Applying cohort-based inclusion criteria
As well as requirements about specific demographics, we may also want to use another cohort for inclusion criteria. Let’s say we want to exclude anyone with a history of cardiac conditions before their Covid-19 cohort entry.
We can first generate this new cohort table with records of cardiac conditions.
cdm$cardiac <- conceptCohort(
cdm = cdm,
conceptSet = list("myocaridal_infarction" = c(317576L, 313217L, 321042L, 4329847L)),
name = "cardiac"
)
cdm$cardiac# Source: table<results.test_cardiac> [?? x 4]
# Database: DuckDB 1.4.0 [unknown@Linux 6.11.0-1018-azure:R 4.4.1//tmp/Rtmp5e21wo/file38b330d37a2c.duckdb]
cohort_definition_id subject_id cohort_start_date cohort_end_date
<int> <int> <date> <date>
1 1 785 2021-02-12 2021-02-12
2 1 1813 1995-08-05 1995-08-05
3 1 2371 2005-07-17 2005-07-17
4 1 3384 2019-07-02 2019-07-02
5 1 3802 2020-10-03 2020-10-03
6 1 3903 2007-01-11 2007-01-11
7 1 4073 1985-01-19 1985-01-19
8 1 7094 1964-02-15 1964-02-15
9 1 7145 1987-02-02 1987-02-02
10 1 7833 1958-04-02 1958-04-02
# ℹ more rowsAnd now we can apply the inclusion criteria that individuals have zero intersections with the table in the time prior to their Covid-19 cohort entry.
cdm$covid <- cdm$covid |>
requireCohortIntersect(targetCohortTable = "cardiac",
indexDate = "cohort_start_date",
window = c(-Inf, -1),
intersections = 0) Note if we had wanted to have required that individuals did have a history of a cardiac condition we would instead have set intersections = c(1, Inf) above.
We could have applied the exact same inclusion criteria using the requireConceptIntersect() function, this code would be equivalent:
cdm$covid <- cdm$covid |>
requireConceptIntersect(
conceptSet = list("myocaridal_infarction" = c(317576L, 313217L, 321042L, 4329847L)),
indexDate = "cohort_start_date",
window = c(-Inf, -1),
intersections = 0
)In fact this approach would be more efficient unless we want to re-use the myocardial_infarction cohort for another inclusion criteria or analysis. Note that the intersection with the cohort table is more flexible as it can have more complicated inclusion/exclusion criteria, but you have to be more careful with the order of inclusion criteria (e.g. if we would restrict the myocaridal_infarction cohort to a certain time span when we would do the intersect we would require to not have the inclusion criteria on that time span).
8.5 Cohort attributes
Using the require*() functions th cohort attribytes have been updated to reflect the applied inclusion criteria.
settings(cdm$covid) |>
glimpse()Rows: 1
Columns: 8
$ cohort_definition_id <int> 1
$ cohort_name <chr> "covid"
$ cdm_version <chr> "5.3"
$ vocabulary_version <chr> "v5.0 22-JUN-22"
$ age_range <chr> "18_64"
$ sex <chr> "Male"
$ min_prior_observation <dbl> 0
$ min_future_observation <dbl> 0cohortCount(cdm$covid) |>
glimpse()Rows: 1
Columns: 3
$ cohort_definition_id <int> 1
$ number_records <int> 158
$ number_subjects <int> 158attrition(cdm$covid) |>
glimpse()Rows: 13
Columns: 7
$ cohort_definition_id <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1
$ number_records <int> 964, 964, 964, 964, 964, 964, 964, 793, 363, 171,…
$ number_subjects <int> 964, 964, 964, 964, 964, 964, 964, 793, 363, 171,…
$ reason_id <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13
$ reason <chr> "Initial qualifying events", "Record in observati…
$ excluded_records <int> 0, 0, 0, 0, 0, 0, 0, 171, 430, 192, 0, 0, 13
$ excluded_subjects <int> 0, 0, 0, 0, 0, 0, 0, 171, 430, 192, 0, 0, 13We can visualise the attrition with the CohortCharacteristics package, we can first extract it with summariseCohortAttrition() and then plotCohortAttrition to better view the impact of applying each inclusion criteria:
attrition_summary <- summariseCohortAttrition(cohort = cdm$covid)
plotCohortAttrition(result = attrition_summary, type = 'png')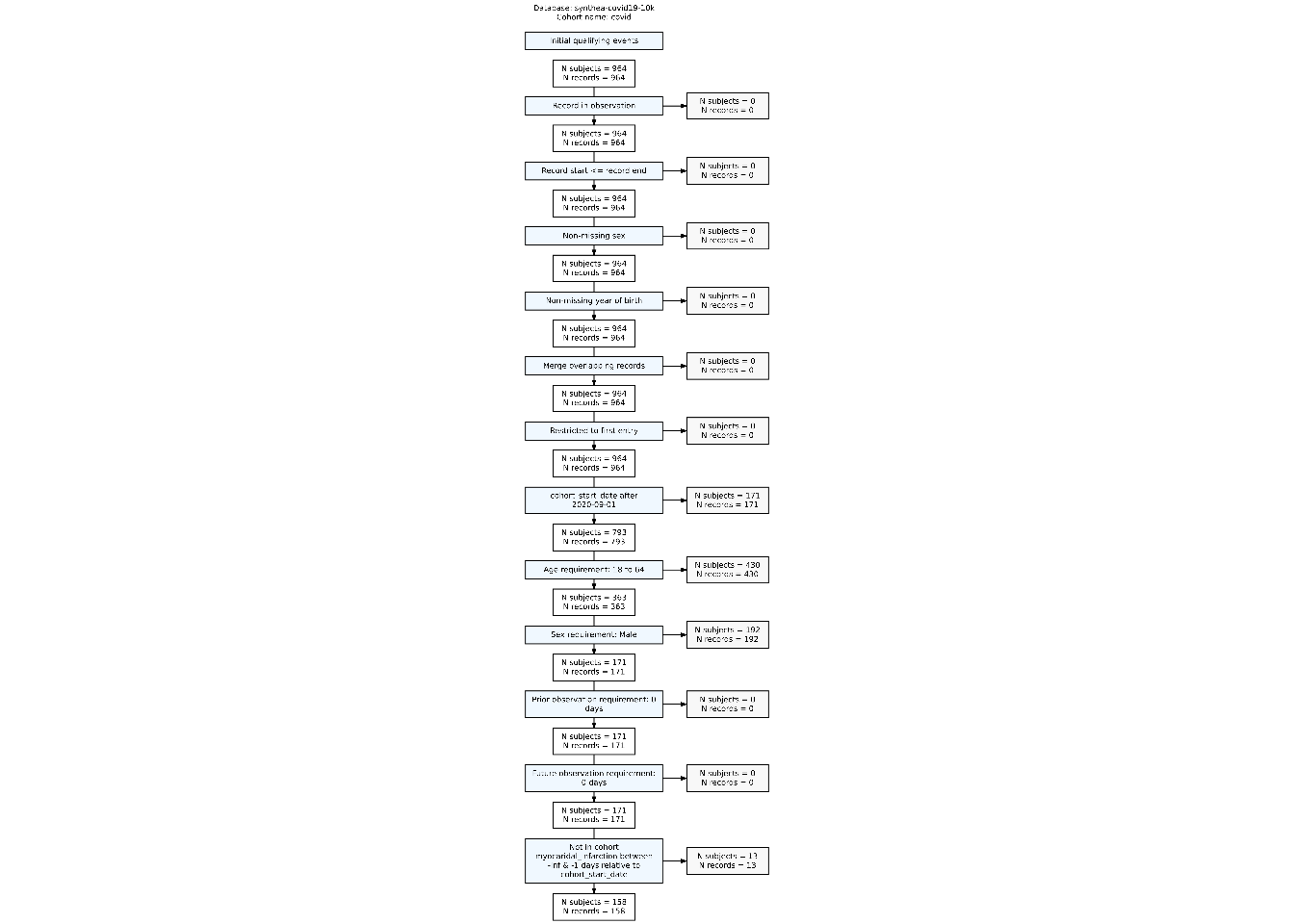
Note that conceptCohort() first step lead to several rows in the attrition table, whereas any other require*() function always add just one record of attrition.
As we have seen that by default the naming of the cohorts in the name of the codelist:
cdm$my_cohort <- conceptCohort(cdm = cdm,
conceptSet = list("concept_1" = 37311061L, "concept_2" = 317576L),
name = "my_cohort")ℹ Subsetting table condition_occurrence using 2 concepts with domain:
condition.
ℹ Combining tables.
ℹ Creating cohort attributes.
ℹ Applying cohort requirements.
ℹ Merging overlapping records.
✔ Cohort my_cohort created.settings(cdm$my_cohort)# A tibble: 2 × 4
cohort_definition_id cohort_name cdm_version vocabulary_version
<int> <chr> <chr> <chr>
1 1 concept_1 5.3 v5.0 22-JUN-22
2 2 concept_2 5.3 v5.0 22-JUN-22 But maybe we are interested to rename a cohort (e.g. after applying the inclusion criteria), we can do that with the renameCohort() utility function:
cdm$my_cohort <- cdm$my_cohort |>
requirePriorObservation(minPriorObservation = 365, cohortId = 1) |>
renameCohort(cohortId = 1, newCohortName = "concept_1_365obs")
settings(cdm$my_cohort)# A tibble: 2 × 5
cohort_definition_id cohort_name cdm_version vocabulary_version
<int> <chr> <chr> <chr>
1 1 concept_1_365obs 5.3 v5.0 22-JUN-22
2 2 concept_2 5.3 v5.0 22-JUN-22
# ℹ 1 more variable: min_prior_observation <dbl>Note that for arguments such as cohortId, targetCohortId, … we are able to use the name of the cohort of interest, see for example:
cdm$my_cohort <- cdm$my_cohort |>
requireSex(sex = "Female", cohortId = "concept_2") |>
renameCohort(cohortId = "concept_2", newCohortName = "concept_2_female")
settings(cdm$my_cohort)# A tibble: 2 × 6
cohort_definition_id cohort_name cdm_version vocabulary_version
<int> <chr> <chr> <chr>
1 1 concept_1_365obs 5.3 v5.0 22-JUN-22
2 2 concept_2_female 5.3 v5.0 22-JUN-22
# ℹ 2 more variables: min_prior_observation <dbl>, sex <chr>This functionality also applies to other packages, such as CohortCharacteristics, PatientProfiles, DrugUtilisation, … Finally in some cases it is useful to add the cohort_name as a column to not have to check manually the equivalence between cohort definition id and cohort name, this can be done using the PatientProfiles utility function addCohortName():
library(PatientProfiles)
cdm$my_cohort |>
addCohortName() |>
glimpse()Rows: ??
Columns: 5
Database: DuckDB 1.4.0 [unknown@Linux 6.11.0-1018-azure:R 4.4.1//tmp/Rtmp5e21wo/file38b330d37a2c.duckdb]
$ cohort_definition_id <int> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2…
$ subject_id <int> 71, 82, 134, 151, 153, 183, 194, 196, 200, 215, 2…
$ cohort_start_date <date> 1936-01-24, 1997-08-04, 2001-01-09, 2022-01-13, …
$ cohort_end_date <date> 1936-01-24, 1997-08-04, 2001-01-09, 2022-01-13, …
$ cohort_name <chr> "concept_2_female", "concept_2_female", "concept_…Also other utility functions that can be useful are those provided by omopgenerics:
library(omopgenerics)
getCohortId(cohort = cdm$my_cohort, cohortName = "concept_2_female")concept_2_female
2 getCohortId(cohort = cdm$my_cohort)concept_1_365obs concept_2_female
1 2 getCohortName(cohort = cdm$my_cohort, cohortId = 1) 1
"concept_1_365obs" getCohortName(cohort = cdm$my_cohort, cohortId = c(2, 1)) 2 1
"concept_2_female" "concept_1_365obs" getCohortName(cohort = cdm$my_cohort) 1 2
"concept_1_365obs" "concept_2_female" 8.6 Disconnecting
Once we have finished our analysis we can close our connection to the database behind our cdm reference.
cdmDisconnect(cdm) 8.7 Further reading
Burn E, Català M, Mercade-Besora N, Alcalde-Herraiz M, Du M, Guo Y, Chen X, Lopez-Guell K, Rowlands E (2025). CohortConstructor: Build and Manipulate Study Cohorts Using a Common Data Model. R package version 0.5.0, https://ohdsi.github.io/CohortConstructor/.