CDM Table name: stem_table (CDM v5.3 / v5.4)
The stem_table is a staging area where HESAPC source codes like Read codes will first be mapped to concept_ids.
Reading from hes_diagnosis_epi
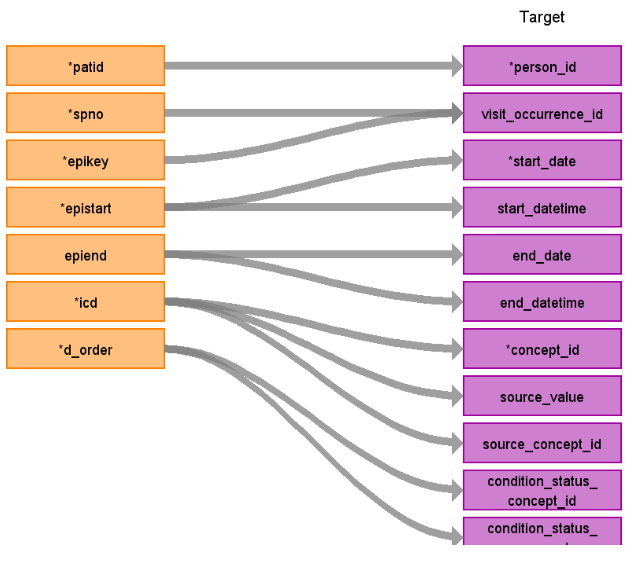
Figure.1
| Destination Field | Source field | Logic | Comment field |
|---|---|---|---|
| id | Removed for performance reasons | ||
| domain_id | NULL | ||
| person_id | patid | ||
| visit_occurrence_id | Use spno to retrieve visit_occurrence_id | ||
| visit_detail_id | Use patid+epikey to retrieve visit_detail_id | ||
| provider_id | NULL | ||
| start_datetime | epistart | ||
| concept_id | icd | If there is no mapping then set to 0 and set domain_id as ‘Observation’.otherwise left join Source_to_Source_vocab_map AS s ON d.ICD=s.SOURCE_CODE AND s.SOURCE_VOCABULARY_ID=’ICD10’ AND target_standard_concept = ‘S’ AND target_invalid_reason is NULL. | |
| source_value | icd | ||
| source_concept_id | icd | concept_id of icd | |
| type_concept_id | 32829 | ||
| operator_concept_id | NULL | ||
| unit_concept_id | NULL | ||
| unit_source_value | NULL | ||
| start_date | epistart | ||
| end_date | epiend | ||
| range_high | NULL | ||
| range_low | NULL | ||
| value_as_number | NULL | ||
| value_as_string | NULL | ||
| value_as_concept_id | NULL | ||
| value_source_value | NULL | ||
| end_datetime | epiend | ||
| verbatim_end_date | NULL | ||
| days_supply | NULL | ||
| dose_unit_source_value | NULL | ||
| lot_number | NULL | ||
| modifier_concept_id | NULL | ||
| modifier_source_value | NULL | ||
| quantity | NULL | ||
| refills | NULL | ||
| route_concept_id | NULL | ||
| route_source_value | NULL | ||
| sig | NULL | ||
| stop_reason | NULL | ||
| unique_device_id | NULL | ||
| anatomic_site_concept_id | NULL | ||
| disease_status_concept_id | NULL | ||
| specimen_source_id | NULL | ||
| anatomic_site_source_value | NULL | ||
| disease_status_source_value | NULL | ||
| condition_status_concept_id | d_order | 32902 if d_order = 1,32908 if d_order > 1 | |
| condition_status_source_value | d_order |
Reading from hes_procedures_epi
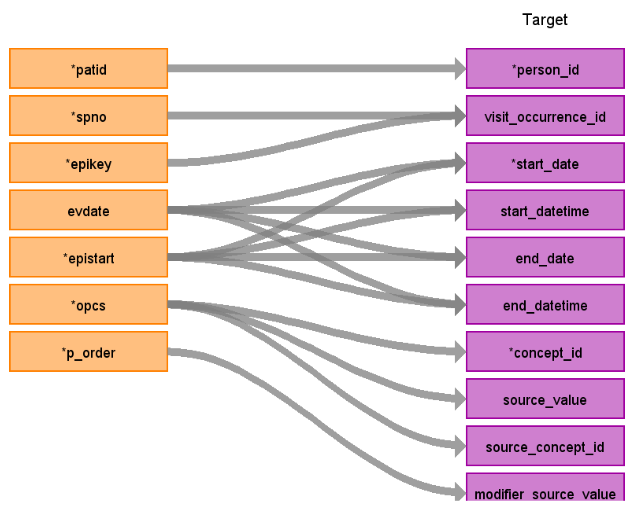
Figure.2
| Destination Field | Source field | Logic | Comment field |
|---|---|---|---|
| id | Removed for performance reasons | ||
| domain_id | NULL | ||
| person_id | patid | ||
| visit_occurrence_id | Use spno to retrieve visit_occurrence_id | ||
| visit_detail_id | Use patid+epikey to retrieve visit_detail_id | ||
| provider_id | NULL | ||
| start_datetime | evdate, epistart | ||
| concept_id | opcs | The codes in the opcs field are four digits with no decimal. To map these. add a decimal between the third and fourth digit. LFFT JOIN Source_to_Source_vocab_map AS ss ON psts.opcs4 = ss.source_code AND ss.source_vocabulary_id = ‘OPCS4’ AND target_standard_concept = ‘S’ AND target_invalid_reason is NULL If domain_id is zero set to ‘Observation’ | |
| source_value | opcs | ||
| source_concept_id | opcs | concept_id of opcs | |
| type_concept_id | 32829 | ||
| operator_concept_id | NULL | ||
| unit_concept_id | NULL | ||
| unit_source_value | NULL | ||
| start_date | evdate | ||
| end_date | evend | ||
| range_high | NULL | ||
| range_low | NULL | ||
| value_as_number | NULL | ||
| value_as_string | NULL | ||
| value_as_concept_id | NULL | ||
| value_source_value | NULL | ||
| end_datetime | NULL | ||
| verbatim_end_date | NULL | ||
| days_supply | NULL | ||
| dose_unit_source_value | NULL | ||
| lot_number | NULL | ||
| modifier_concept_id | NULL | ||
| modifier_source_value | p_order | ||
| quantity | NULL | ||
| refills | NULL | ||
| route_concept_id | NULL | ||
| route_source_value | NULL | ||
| sig | NULL | ||
| stop_reason | NULL | ||
| unique_device_id | NULL | ||
| anatomic_site_concept_id | NULL | ||
| disease_status_concept_id | NULL | ||
| specimen_source_id | NULL | ||
| anatomic_site_source_value | NULL | ||
| disease_status_source_value | NULL | ||
| condition_status_concept_id | NULL | ||
| condition_status_source_value | NULL |